- Laboratory
- Molecular biology
- Analysis software module
- JSI medical systems
NGS sequencing software module SEQNEXTanalysiscapturefor mapping
Add to favorites
Compare this product
fo_shop_gate_exact_title
Characteristics
- Function
- analysis, capture, for mapping
- Applications
- laboratory, for NGS sequencing
Description
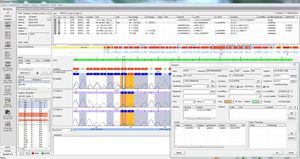
SEQNEXT is a powerful and user-friendly application for mapping, alignment and variant detection of Next Generation Sequencing data. SEQNEXT can analyse data from all common sequencing platforms and kits (PCR-based, enrichment, sequence capture). Visualisation of all detected variants like deletions, insertions and indels of any length, SNPs, repeat regions as well as CNVs and gene fusions is clear and intuitive.
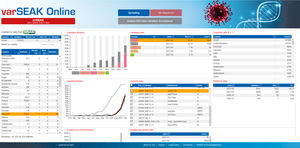
SEQNEXT offers access to public SNP-databases like dbSNP, 1000 Genomes, ClinVar, ExAC and gnomAD for classification and filtering. All result data can be exchanged with our variant database for Shared Experience And Knowledge - varSEAK, transferred to laboratory internal LIM Systems and / or issued as personalised patient reports.
NEW Improved Analysis of Virus sequencing data e.g. SARS-CoV-2
Trioanalysis and improved Poolanalysis
Sequence Ontology Annotations in the variant table
Jumping option to varSEAK and all available external DBs like dbSNP, ClinVar, UK10K, ESP, gnomAD and ExAC
Whole Exome Sequencing (WES) analysis
Compatible with data from all common Next Generation Sequencing platforms and kits
Easy set up of individual target regions (ROIs) via import of bed- or manifest-files based on hg19 and / or hg38
Import of gene lists for filtering and analysis of WES data
Analysis of fastq-, fastq.gz- or bam-files
Efficient standards for mapping, alignment, quality and variant calling (used algorithms: BWA and Smith-Waterman, adapted by JSI)
Personalized settings for analysis and variant calling (≥ 0.1 %)
High sensitivity and specificity for detection of deletions, insertions and indels of any length, SNPs, repeat regions as well as CNVs and gene fusions
Catalogs
SEQUENCE PILOT
2 Pages
*Prices are pre-tax. They exclude delivery charges and customs duties and do not include additional charges for installation or activation options. Prices are indicative only and may vary by country, with changes to the cost of raw materials and exchange rates.