- Laboratory
- Molecular biology
- Analysis software module
- JSI medical systems
HLA sequencing software module SEQHLAanalysislaboratory
Add to favorites
Compare this product
Characteristics
- Function
- analysis
- Applications
- laboratory, for HLA sequencing
Description
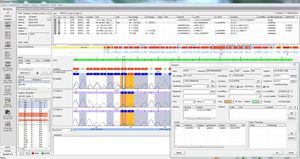
SEQHLA is a powerful and user-friendly application for the analysis of HLA sequence based typing data from all Sanger sequencing platforms. All IMGT HLA database versions are available for download and import. It uses an unique learning base caller based on peak area statistic data and has its own auto edit function.
Constant positions can be ignored for analysis if desired. The result can be displayed in maximum, 1 field or 2 field resolution and as NMPD code. All result data can be transferred to laboratory internal LIM Systems and / or issued as personalised patient reports.
Compatible with data from all common Sanger sequencing platforms and kits
Easy setup of individual target regions based on the IMGT HLA database
Configurable base caller with sequencer dependent thresholds
Automatic and user defined trimming of sequencing result files
All results for one patient shown in one screen incl. detected allele combinations
Automatic warning for splice site mutations
Possibility of ignoring constant positions for the analysis
Detection of new alleles
Display of results in different resolutions (1 field, 2-field, and max. resolution) and as NMDP code
Peak area statistic: an automatic comparison to all previously analysed samples for a sensitive detection of heterozygous positions even in case of an allelic dropout
Detailed HLA report for individual patients / orders
Catalogs
SEQUENCE PILOT
2 Pages
*Prices are pre-tax. They exclude delivery charges and customs duties and do not include additional charges for installation or activation options. Prices are indicative only and may vary by country, with changes to the cost of raw materials and exchange rates.