- Laboratory
- Laboratory medicine
- NGS sequencing software
- PhenoSystems SA

- Products
- Catalogs
- News & Trends
- Exhibitions
NGS sequencing software GensearchNGSlaboratoryanalysisdata management
Add to favorites
Compare this product
Characteristics
- Applications
- for NGS sequencing, laboratory
- Function
- analysis, data management, diagnostic
Description
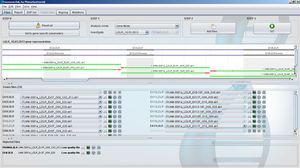
GensearchNGS is an integrated software solution for the analysis of DNA-Seq data from commonly used NGS equipments such as Roche/454, Illumina, Ion Torrent and Proton. It has been designed and developed in close collaboration with leading accredited diagnostics groups in Europe and with the support of the NMDchip FP7 project.
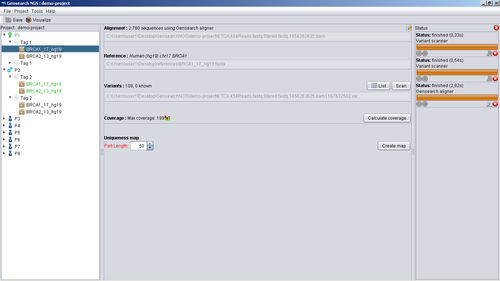
It integrates all the steps from importing and filtering sequence reads, bar code splitting, alignment, variant detection and annotation to the final report of detected variants. Variants can be annotated with various public and private data sources (Ensembl, Alamut, Genome Trax,...) They can be filtered by logical combinations between samples for family studies.
Main features:
User friendly software for resequencing NGS projects in a diagnostic setting, running on Windows, Linux and MacOS systems.
Connects*) to variant interpretation tools, and databases such as: Ensembl, dbSNP, Alamut, Clinvar, Wiki Pathways,Gene Ontology*).
Filter variants across samples (e.g. variants common to several samples or exclude variants of one sample from another).
Patient centric database system to manage sequence data and validated variants.
Customisation possible to export data into your laboratory management system.
Possibility to publish your variants on Clinvar*).
Developed together with accredited laboratories.
For larger projects (WES/WGS): possibility to use multiple PCs in parallel.
Plugin technology: alignment with our proprietary or public algorithms such as Bowtie, BWA or Stampy.
Annotation and filtering with HPO, OMIM and Orphanet descriptors and codes, helpful when fishing for mutations in whole exomes or whole genomes.
*Prices are pre-tax. They exclude delivery charges and customs duties and do not include additional charges for installation or activation options. Prices are indicative only and may vary by country, with changes to the cost of raw materials and exchange rates.