- Medical Technical Facilities
- Healthcare IT, Telemedicine
- Analysis software
- SoftGenetics, LLC.
Analysis software GeneMarker®HTS for controlreportingNGS

Add to favorites
Compare this product
fo_shop_gate_exact_title
Characteristics
- Function
- analysis, for control, reporting, NGS
- Applications
- medical, for DNA sequencing
Description
GeneMarker®HTS software provides a validated streamlined workflow for forensic mitochondrial , STR, and Y-STR casework as well as medical research of mitochondrial DNA from massively parallel squencing platforms (MPS) - such as the Illumina® and Ion Torrent® - in an easy-to-use Windows® operating system.
GeneMarkerHTS (High Through-put Sequence) Forensic Analysis Software for data from Next Generation Sequence (NGS), Massively Parallel Sequencing (MPS) platforms, including Illumina® and Ion Torrent®
Mitochondrial DNA Analysis
Whole Genome, HVI/HVII and control region analysis
Unique Alignment Technology ∙ Motif Consensus
Forensic Nomenclature
Easy Upload to EMPOP
STR Analysis
Autosomal & Y-STR
Forensic Nomenclature
Genotype & SNP Reporting
Concordant
Validated Easy-to-Use Windows® Interface
Compatible with major Chemistries & Platforms
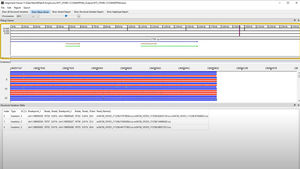
Audit Trail & User Control
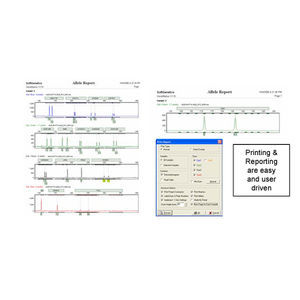
Comprehensive Reporting Options
GeneMarkerHTS software includes:
Audit trail capability
User management
Customizable viewing and reporting to protect privacy of personal health information (PHI)
Profile Comparison Capabilities
Upload to EMPOP
Rapid Analysis – get results in minutes
mtDNA:
30 MiSeq whole mtDNA chromosome data files with 10,000 average depth of coverage were aligned in 90 minutes (3 minutes per sample).
200 whole mtDNA chromosome data files with 10,000 average depth of coverage aligned in 16 hours.
STR/Y-STR
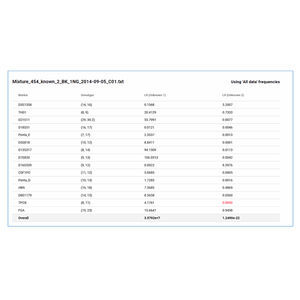
GeneMarker HTS software results were 99.74% concordant with the CE allele calls of 20,000 sampled GeneMarker loci.
Samples allele calls and iso-alleles
VIDEO
Catalogs
No catalogs are available for this product.
See all of SoftGenetics, LLC.‘s catalogs*Prices are pre-tax. They exclude delivery charges and customs duties and do not include additional charges for installation or activation options. Prices are indicative only and may vary by country, with changes to the cost of raw materials and exchange rates.